End-member modelling analysis (EMMA) – unmixing grain-size data to decipher process systems
The grain-size distributions of sediment deposits routed across the Earth surface contain valuable information about the source area they are derived from, the transport processes that moved them and the transport pathway. These information about the transport regime are however obscured when sediments become mixed during deposition. End-member modelling analysis or EMMA is a statistical approach to unmix the underlying transport regimes.
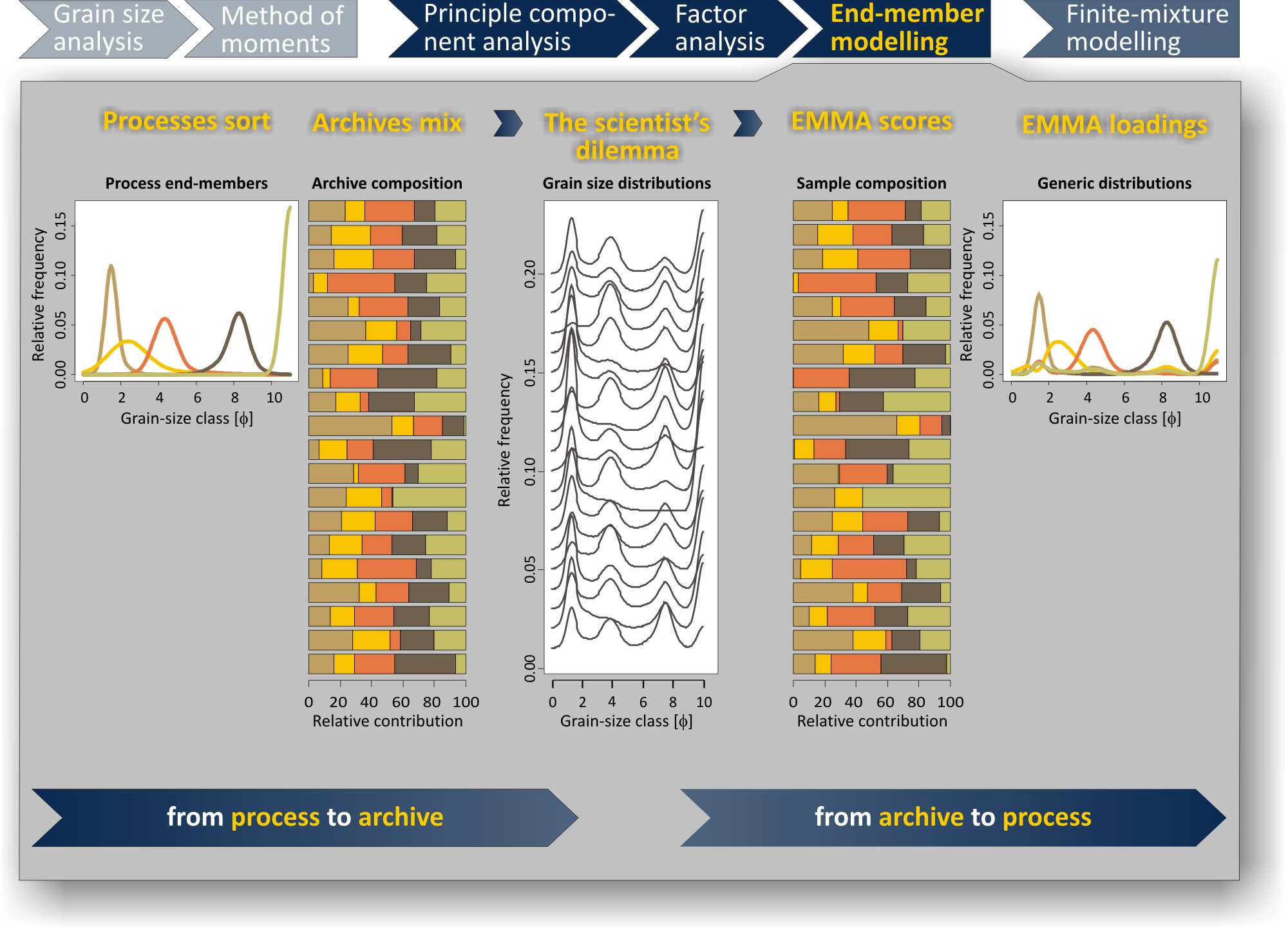 The figure above explains the principal setting of EMMA. From grain-size measurements classic statistic parameters based on the method of moments (e.g., mean, skewness, D50) can be calculated but are only meaningful for unimodal distributions. As soon as the samples consist of more than one unimodal distributions these need to be unmixed, either statistically (by EMMA) or by fitting parameteric models (e.g., the sum of log-normal distributions).
The figure above explains the principal setting of EMMA. From grain-size measurements classic statistic parameters based on the method of moments (e.g., mean, skewness, D50) can be calculated but are only meaningful for unimodal distributions. As soon as the samples consist of more than one unimodal distributions these need to be unmixed, either statistically (by EMMA) or by fitting parameteric models (e.g., the sum of log-normal distributions).
The underlying concept of EMMA is that processes tend to sort sediments and create characteristic grain-size distributions. When several processes deposit sediments at a given place (a lake, an ocean, a loess deposit) the process signatures become mixed. EMMA describes such a mixed data set as a combination of end-member loadings and scores. Loadings are the fundamental grain-size distributions that build the data set and may be interpreted as a direct proxy for the responsible processes. Scores are the relative contributions of each loading to a sample and may be interpreted as the importance of each process in the formation of the deposit through time or space.
WHO BUILT EMMA?
The algorithm that is the heart of the R-package EMMAgeo was developed for Matlab by Elisabeth Dietze (references see below). I simply translated it to R and built a couple of functions around it to allow a comprehensive and convenient workflow in a free and open software environment. If you have questions about the EMMA algorithm, its concept and mathematical foundations, please contact Elisabeth directly. If you request technical support and need information about the functions of the R-package, see below and feel free to contact me.
HOW TO CITE THE PACKAGE
The reference article on EMMAgeo is now published! The text that gives a user perspective on EMMAgeo along with a comparison of EMMA with other unmixing approaches as well as templates for the full workflow in EMMA is available as Open Access article here. If you want to use EMMAgeo in your work please use the following two (yes both) references.
Dietze E and Dietze M. 2019. Grain-size distribution unmixing using the R package EMMAgeo, E&G Quaternary Sci. J., 68, 29–46, https://doi.org/10.5194/egqsj-68-29-2019.
Dietze E, Hartmann K, Diekmann B, IJmker J, Lehmkuhl F, Opitz S, Stauch G, Wünnemann B, Borchers A. 2012. An end-member algorithm for deciphering modern detrital processes from lake sediments of Lake Donggi Cona, NE Tibetan Plateau, China. Sedimentary Geology 243-244: 169-180.
GETTING THE PACKAGE
EMMAgeo is hosted on CRAN, the comprehensive R archive network. Thus, it is easy to install using
install.packages("EMMAgeo").
Currently, version 0.9.7 is online. The latest developer version with mostly minor additions and bug fixes is hosted on GitHub. There are a series of ways to install the package. The most convenient and coherent one is installing it through the R-package devtools.
devtools::install_github(repo = "coffeemuggler/EMMAgeo", ref = "dev_0.9.8")
Of course, this requires installing devtools first (e.g., by typing install.packages("devtools")). If you need help installing devtools, see the help page on the CRAN website. Windows users, in order to install the package devtools, you need to install the software RTools, first.
Alternatively, if you do not want or do not manage to work with devtools these versions can be downloaded (no guarantee for the very latest version, see time stamp on GitHub) and used for installation typing
install.packages(pkgs = "~/Downloads/EMMAgeo_0.9.8.tar.gz", repos = NULL, type = "source")
where "~/Downloads/EMMAgeo_0.9.8.tar.gz" is the location where you downloaded the package source archive.
| Linux/Mac OS | EMMAgeo_0.9.8.tar.gz | compiled 2019-12-17 |
| Windows OS | EMMAgeo_0.9.8.zip | compiled 2019-12-17 |
EMMAgeo has seen quite a bit of success and wide application. Hence, there is material of differnt depth and scope available. Currently, we are working on a profound validation article.
- A brief interactive PICO presentation
- Slides of a workshop at University Cologne
- Don't forget the package documentation and the links below.
GALLERY OF EMMA APPLICATIONS
- EMMA untangles evolution of permafrost sediments.
- EMMA unmixes environmental drivers in seismic records, Example.
- EMMA quantifies processes of hillslope cover bed sediment formation.
- EMMA unmixes deposits under stone pavements in the Mojave Desert, Dietze et al. (2016)
- EMMA identifies marine dynamics continuosly in laminated sediments, Example.
- EMMA reveals coherent sedimentation patterns across Tibetan lakes, Dietze et al. (2014)
- EMMA sheds light on sedimentation dynamics in a Jurassic deltaic system.
- EMMA quantifies sedimentation patterns in the Nile delta
BUGS AND REQUESTS
If you feel helpless with an issue or notice a bug in one of the functions of the package, if you wish to have a further feature implemented, there is a good chance that I will engange with this issue, if you let me know of it. Please send me an email and I will add this to the list of issues shown here.
Bug in GUI(). Example data set not found. GUI cannot start
Bug fixed. Example data set name updated according to latest version.
Bug in robust.EM(). Classunits are not shown in legend, even if provided.
Bug fixed. Classunits are now plotted.
Add option to get.l.opt() to find l_opt automatically by allowing the keyword l = "opt".
Add function fit.em() to fit parametric models to clipped end-members.
Add function clip.em() to extract only main modes of end-members until the graph is no longer falling to lower values.
In EMMA(), the arguments lty and lwd are only supported n the legend, not in the actual plot part. Manual labels cannot be set for Rn-part of the plot.
Just as an idea, write function validate.model() that creates random mixing ratios and builds an artificial data set with EMMA output (Vqsn), unmixes it and provides measures of model quality.
Is it possible to also deliver measures of uncertainty for R and E output of robust EMMA?
Catch error and function stop of robust.EM() when no end-members are found with the given limits.
Add argument "repair = TRUE" to test.parameters().
Allow legend in images of test.parameters(), like in fields::image.plot().